The rapid progress in computer vision has enabled automation and assistance in a variety of fields. The medtech industry is no exception. Data-driven segmentation models have already reduced the time radiologists spend on manual annotations [1].
But as in every machine learning task, data is the bottleneck. Annotated medical images are expensive and time-consuming to produce [2], and data often sits locked inside isolated silos at different hospitals and clinics.
Federated Learning (FL) has emerged as a way to train models across multiple data silos without sharing raw data. It enables collaboration while maintaining patient privacy [3].
However, FL doesn’t fully solve the segmentation challenge. Annotation standards differ between hospitals, and even between radiologists in the same clinic. This lack of consistency makes pooling labeled segmentation data less effective than we might hope. This was one key learning in the ASSIST project [4].
For related image analysis tasks in automotive, we have recently demonstrated the potential of federated learning together with self-supervised learning. The same strategy can also unlock silos in medical imaging, which is what we explore in this post.
Manual annotation is a blocker to scale modeling across data silos. This is where Self-Supervised Learning (SSL) can make a difference. Unlike supervised training, SSL can leverage all available data, even if it’s not annotated.
Recent methods like DINO [5] and DINOv2 [6] have shown that Vision Transformers (ViTs) can be pre-trained on massive amounts of unlabelled images to learn strong, general-purpose representations. Once such a backbone is trained, it can be fine-tuned for a specific downstream task, such as medical image segmentation, using only a small set of annotated examples.
In other words: SSL makes it possible to extract value from isolated medical datasets without needing consistent, large-scale annotation.
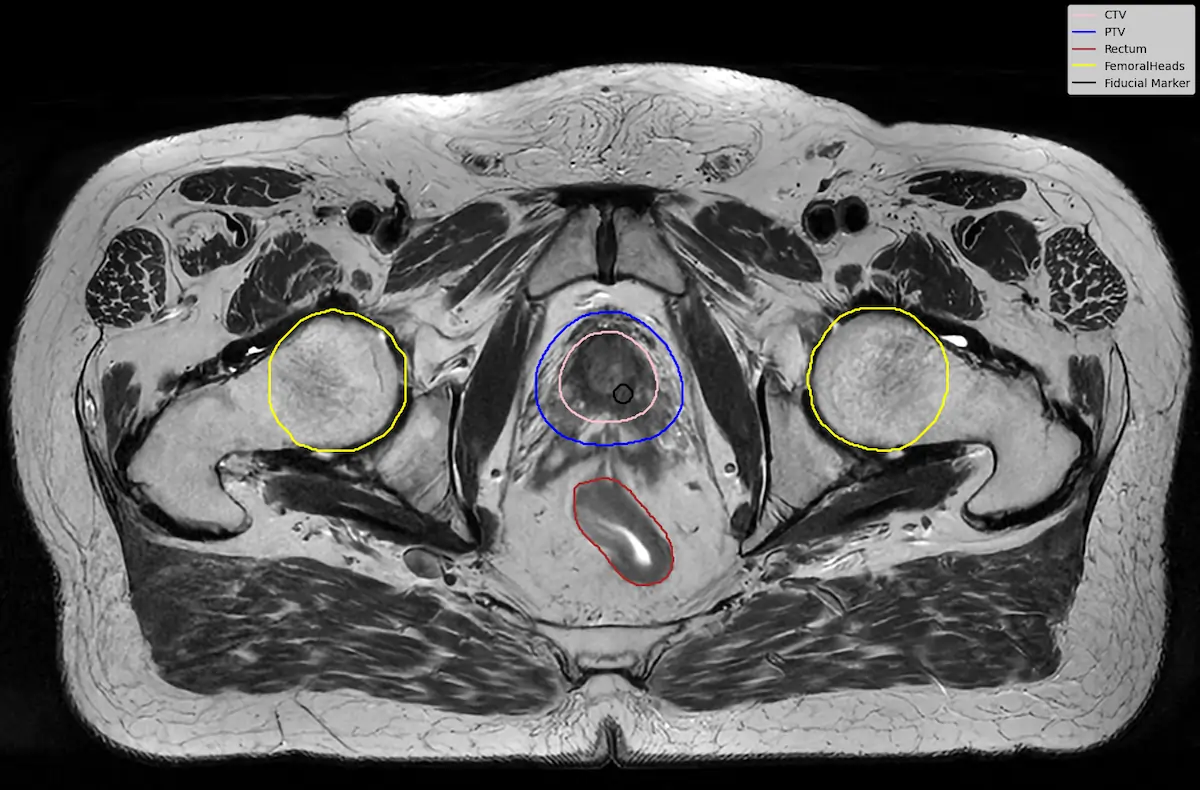
To test this idea, we performed a small downstream experiment using the LUND-PROBE dataset from Lund University [7]. The dataset contains multiparametric prostate MRI scans collected as part of the PROBE study, aimed at developing AI methods for improved prostate cancer detection and diagnosis. The data is de-identified and curated for research purposes, making it a valuable benchmark for medical imaging tasks.
Our best SSL-based model slightly outperformed the traditional U-Net baseline, even with limited annotated data. This shows SSL can match or beat established methods using fewer annotations.
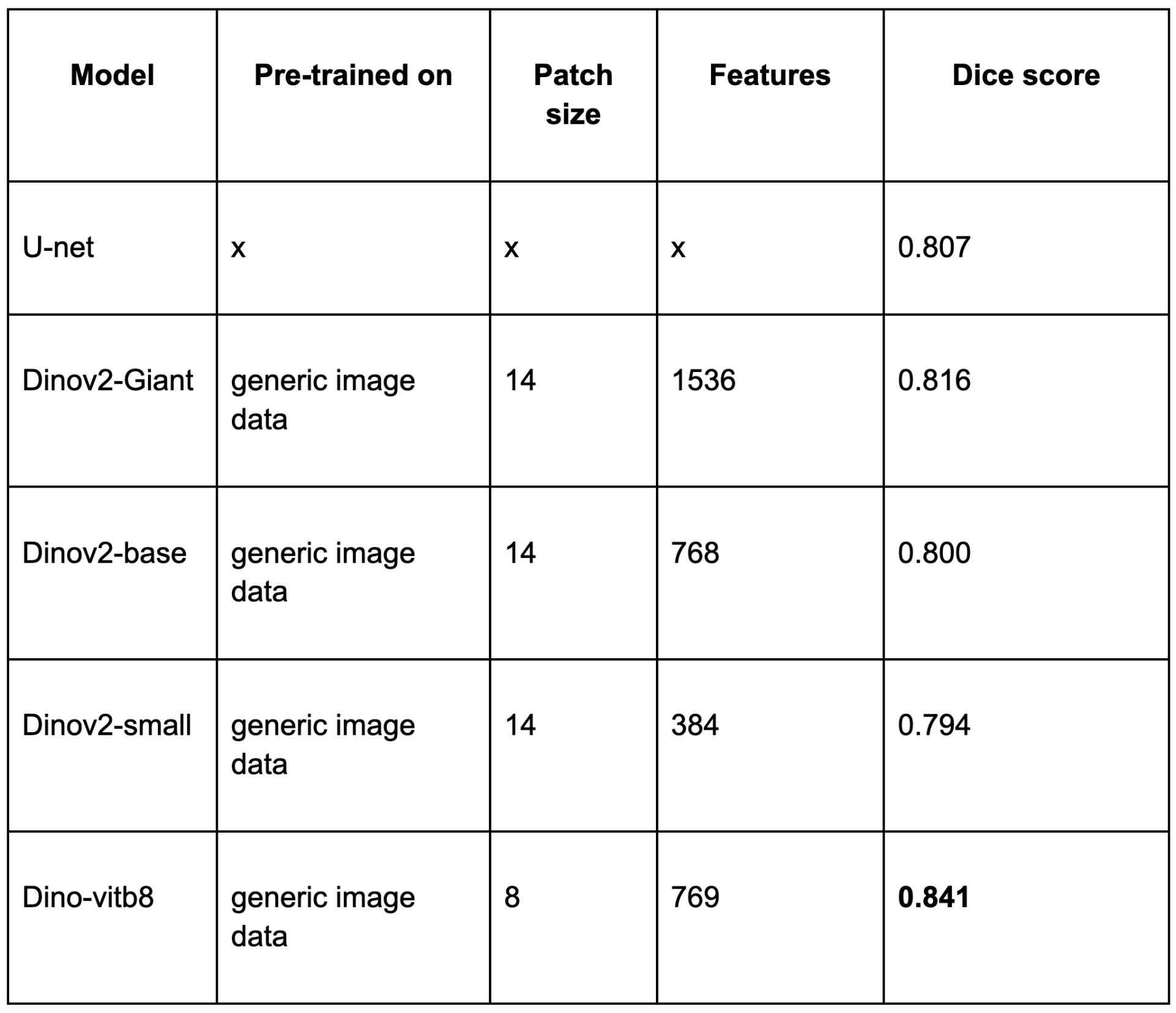
These findings highlight a few important points:
Self-Supervised Learning is not just a technical trick, it’s a new way of approaching the problem. For medtech, it offers a way to turn the challenge of isolated, inconsistently labeled data into an opportunity. By first training strong backbones on unannotated data and then fine-tuning them on small, local datasets, we can build models that are both powerful and practical.
For clinicians and researchers, this means faster AI development with fewer bottlenecks. For patients, it means faster access to better diagnostic tools.
The next step? Exploring domain-specific SSL pre-training across medical silos, federated and annotation-free. An exciting future direction is also the design of 3D-ViTs, enabling the models to fully leverage the spatial information in volumetric medical images.
If you are interested in experimenting with federated DINO training, take a look at Scaleout’s VisionKit package, which integrates seamlessly with Scaleout Edge and includes federated implementations of both DINO and DINOv2.
Author Mattias Åkesson, Senior Federated Learning Engineer
---
[1] Tajbakhsh, N., et al. (2020). "Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation." Medical Image Analysis.
[2] Lundervold, A. & Lundervold, A. (2019). "An overview of deep learning in medical imaging focusing on MRI." Zeitschrift für Medizinische Physik.
[3] Sheller, M.J., et al. (2020). "Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data." Scientific Reports.
[4] ITEA4 Project ASSIST. Available at: https://itea4.org/project/assist.html
[5] Caron, M., et al. (2021). "Emerging properties in self-supervised vision transformers." ICCV.
[6] Oquab, M., et al. (2023). "DINOv2: Learning robust visual features without supervision." arXiv:2304.07193.
[7] LUND-PROBE dataset, Lund University. Available at: https://datahub.aida.scilifelab.se/10.23698/aida/lund-probe